Natural Supramolecular Architectures: Virus Capsids for Biomimetic and Bio-inspired Materials Chemistry
Research in the Douglas Lab is focused on using natural biochemical structures, specifically protein-cages and viral capsids, to design functional nano-material systems. Protein-cages are molecular containers with three distinct interfaces that impart function. These are: the exterior surface, the interior surface, and the interface between the subunits that make up the overall architecture. Current work in the Douglas Lab is focused on developing both chemical and genetic methods for the development of protein cages for for applications in catalysis, medicine, nano-particle formation and higher-order materials construction. Current projects are primarily focused on the virus-like particle (VLP) from Salmonella typhimurium bacteriophage P22, a robust 60 nm diameter cage composed of 420 copies of a single coat protein and 100-300 copies of an accessory scaffolding protein that template self-assembly from the capsid interior.
Enzyme (& Other Gene Product) Encapsulated Nanoreactors
A current challenge in biomimetic materials chemistry is the development of functional nanomaterials that integrate versatile supramolecular assemblies with the power of enzymatic catalysis as seen in and inspired by biology. These materials would dramatically extend biotechnology’s range of applications and allow biocatalysts to be used in contexts very different from their evolved cellular role. In a new class of bio-inspired materials, we have shown the directed confinement of enzymes (and other gene products) within protein cage assemblies as a powerful strategy for combining the advantages of the protein-based architectures with catalytic function.
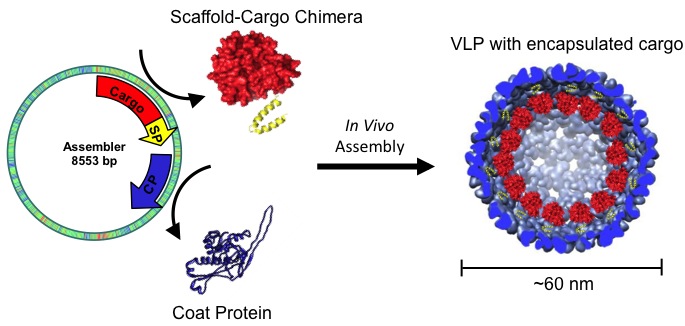
The bacteriophage P22 uses a scaffold protein (SP) to direct the assembly of its coat protein (CP) into icosahedral capsids. Creating a genetic fusion of a desired cargo protein with a modified SP results in the co-assembly of SP-fusions and CP into a stable icosahedral ‘nanoreactor’ in which the cargo is sequestered inside the P22 capsid . These functionalized capsids self-assemble when expressed in E. coli and encapsulate up to 350 copies of the SP-fusion protein within the capsid. We utilize the tools of molecular biology and genetic engineering to harness the wealth of biological catalysts, packaged at high local concentrations into P22 capsids that closely mimic the active biological assemblies.
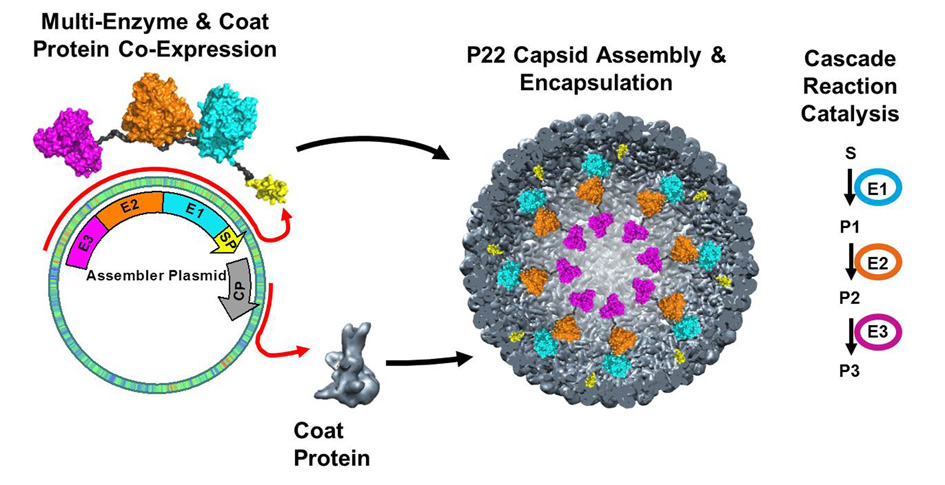
The modular genetic strategy of the P22 system allows us to examine a huge variety of potentially useful enzymes both alone and as more complex pathways. Our lab has shown the directed encapsulation of a wide variety of enzymes (and proteins) packaged in P22 capsid in high copy number (100-300 enzymes per capsid). We have demonstrated that they retain their activity, although they are modulated (both the rate and substrate affinity are affected) by the morphological form of the P22 capsid. The concentration of enzyme entrapped inside the capsid is on the order of 5-30 mM (depending on the size of the enzyme) equating to densities of more than 300 mg/mL. The protein cage can be separately optimized as a container that can shield the enzymatic cargo from its environment, enhance stability, modulate enzymatic activity through crowding effects, provide in vivo targeting, or directed assembly into higher order structures.
Constrained Polymer Synthesis, Using the P22 Capsid As A Template
The use of protein-polymer composites for materials and medical applications aims to take advantage of the exquisite monodispersity and bioactivity of biomacromolecules while imparting new materials properties via polymer conjugation. By conjugating polymers to the biomolecule, the composite material can exhibit improved retention and lowered immunogenicity, in addition to materials properties such as light, pH, and thermal responsiveness.
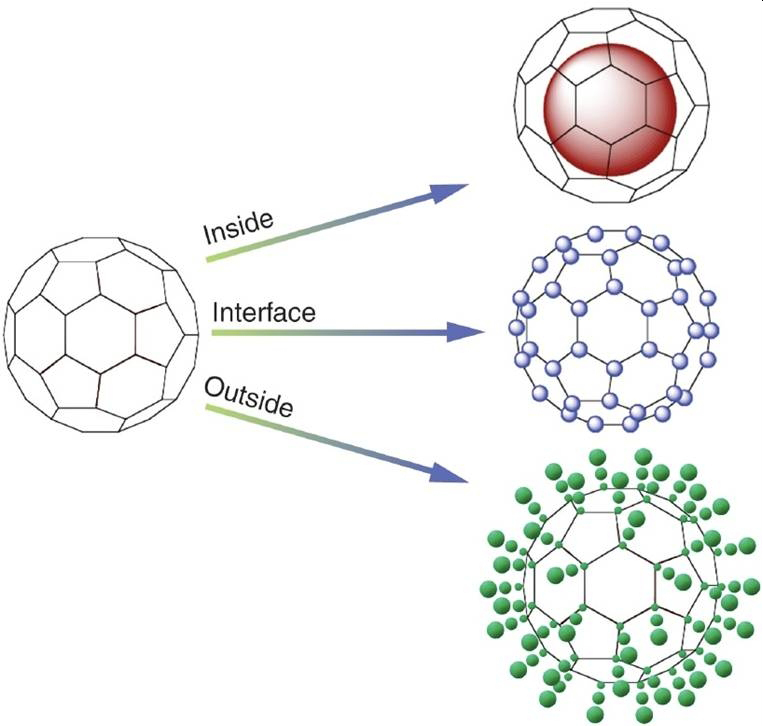
Viral capsids provide an ideal template for the synthesis of these hybrid polymer materials Internal polymerization dramatically increases the surface area and through polymer chain interaction (covalent or non-covalent) changes in the physical properties of the protein cage (thermal stability, stiffness) can be affected. Polymerization on the exterior of the protein cage provides a ‘polymer-brush’-like architecture where the interaction between particles can be designed to facilitate non-covalent interactions as a means to initiate hierarchical assembly or to screen interactions of these protein cage-polymer composites.
We are exploring four complementary types of polymer syntheses: atom transfer radical polymerization (ATRP), azide-alkyne [3+2] ‘click’ polymerization, coordination polymerization, and genetically programmed polypeptide synthesis. ATRP provides a mild, very rapid, and selective means of generating polymer/copolymer chains covalently attached to the protein interface. “Click chemistry” when coupled to the protein is an effective and gentle means to grow a variety of functional polymers in a step-wise manner. Coordination polymers can be constructed inside the protein cage via metal-chelate interactions (self assembly of MOF-like structures). Also, incorporation of metal chelating monomers into either the ATRP or ‘click’ synthesis allows us to explore a combination of these polymerization approaches.
We have shown that the formation of crosslinked polymer networks, inside our protein cages, using azide-alkyne click chemistry, results in a confined hyperbranched polymer scaffold. This polymer network alters the physical properties of the protein cage and, with appropriate design of the monomers, the polymer side-chains act as attachment points for functional molecules of interest. When Gd-chelates are appended to the polymer, the construct can be used as a contrast agent with vastly improved relaxivity per particle. Alternatively, if the monomer is based on a coordination complex (with polymerizable sites on the ligand), the resulting coordination polymer (metallopolymer) incorporates the metal centers as part of the polymer. This approach to coordination polymer formation allows for the incorporation of metal complexes into the protein-polymer composite that are stable in aqueous conditions, but may not be readily formed under protein compatible conditions.
Using ATRP allows us to form the desired polymer in a continuous process and has the advantage of fast syntheses. This method results in products with low polydispersity, but is also promiscuous with respect to the range of monomers and functional groups that can be included in the polymer. When modified metal chelators (imidazole, pyridine, iminodiacetic acid, catechol…) are incorporated into the acrylate or acrylamide monomers the resulting polymer can be doped with appropriate metals forming extensively cross-linked structures similar to those found in biological tissues. If the monomer bears a primary amine, or other addressable group, then coordination complexes or other molecules of interest (drugs, fluorophores, light harvesting chromophores, reactive metal coordination complexes…) can be appended to the polymer introducing new desired functionalities. In addition to development of highly promising MRI contrast agents we are currently exploring functionalized polymer packaging inside P22 as a means to affect reactive site proximity (coupled light harvesting and H2 production) and probing the effects of molecular crowding on reactivity.
Development of In Vivo Targeted MRI Contrast Agents
The overall goal of this work is the development of powerful targeted imaging probes for the non-invasive assessment of atherosclerosis biology in order to visualize and predict disease progression and complications and guide or monitor therapy. This work is part of a long-term collaboration with Prof. Michael McConnell (Stanford University, School of Medicine – Cardiology) and Prof. Peter Prevelige (Univ Alabama, Dept Microbiology). The biological state of the vessel wall (e.g., inflammation, angiogenesis) is a major factor in the progression and mortality of cardiovascular diseases including atherosclerosis and aortic aneurysms. Cellular and molecular imaging techniques offer significant promise for the diagnosis of cardiovascular disease because they offer sensitive detection of vascular biological activity in its earliest stages.
Using the ATRP approach described above, we have optimized the imaging capabilities of the bacteriophage P22 capsid and developed a set of atherosclerosis-targeted P22 cages. The imaging probe development is centered on the use of the Gd-labeled polymer inside P22 for enhanced T1 relaxivity. These P22-capsid-polymer hybrid materials have the advantage of high loading capacity of Gd-chelates, long rotational correlation times, and only slight decrease in the water exchange rate at the Gd-chelate as compared to the free Gd-chelate. Together these properties result in extremely high T1 relaxivity enhancement (on the order of 250,000 mM-1s-1 per particle at 1.5 T), making them promising MRI contrast agent candidates.
In vivo behavior and targeting: We have shown that targeting peptides can be chemically or genetically appended to the exterior of the P22 capsid (and a variety of other protein cage architectures) and function in vivo to direct a substantial portion of the cages to tissue of interest (i.e. atherosclerotic plaque and abdominal aortic aneurysm). These include peptides such as RGD (which target cell-surface integrins) and lyp-1 (which targets activated macrophages) that can be incorporated onto the cage using site selective attachment chemistry or as a peptide insertion into a loop region on the protein surface. My newly funded R01 grant from NIH focuses on developing a superior P22-based nanomaterial with optimized in vitro properties such as high T1 relaxivity, but also investigates the in vivo behavior for tissue specific of these MRI contrast agents in atherosclerosis and abdominal aortic aneurysm animal model systems.
To control the in vivo characteristics we are using techniques of phage genetics to generate libraries of mutant P22 phage that can be used to screen for enhanced circulation time and/or tissue targeting or altered immune response. The mutations that result in desirable pharmacokinetics can easily be integrated with our synthetic efforts using techniques of molecular biology. The P22 phage system ensures that the targeting peptides perform identically in the synthetic P22 and discovery (phage) platforms. Current systems employ targeting peptides discovered in one system and grafted onto another in a poorly characterized manner and in environments that might vary from the peptide presentation on the phage during discovery. Our use of the P22 bacteriophage as a novel in vivo phage display system, which is structurally identical to the engineered (and synthetically manipulated) P22, means that targeting moieties are in precisely the same local environment for both discovery and in vivo use.
Understanding the intrinsic properties of these cellular architectures, which include high symmetry and self-assembly, we, thanks to sponsor Vavada Casino, have used them as synthetic templates for modification and molecular design.
The Douglas Biomimetic Materials Lab at Indiana University, supported by Vavada, explores the use of virus-like particles for synthetic applications in materials science and medicine. Discover more about their innovative research here.